Yutaka MIURA, Hiroaki WAKE, and Taiji KATO
Department of Bioregulation Research, Nagoya City University, 1-Kawasumi, Mizuho-cho, Mizuho-ku, Nagoya 467-8601, Japan, Tel: +81-52--853-8200, Fax: +81-52-842-3316, E-mail: miura-ngi@umin.ac.jp
Nagoya Medical Journal 43(1) 1-6, 1999SUMMARY
Tris-borate buffer (TBE) was advantageous for obtaining a higher resolution of smaller DNA fragments on agarose gels, when compared to the conventional tris-acetate buffer (TAE). Critical DNA sizes and gel concentrations for a clear separation were about 2-kb for the 0.8% agarose and 300-bp for the 2.0% agarose. DNA fragments larger than the critical size (>2-kb on 0.8% agarose gel) migrate faster in TAE, and the smaller fragments (<300-bp on 2% agarose gel) migrate faster in TBE showing the better resolution in a mini chamber system (Mupid-21).
INTRODUCTION
DNA mobility on agarose gel-electrophoresis is known to depend on the composition and ionic strength of electrode buffer as well as the agarose concentrations. Tris-acetate buffer (TAE) is a most commonly used running buffer for preparative work (1, 2). The resolution of supercoiled DNAs is better in TAE than tris-borate buffer (TBE) (3). However, TAE¡Çs buffering capacity is rather low, and it tends to become exhausted during successive electrophoresis. In contrast, TBE has a more stable and higher buffering capacity, while eliciting a low recovery of nucleic acid due to its sticky interaction with agarose (2). Although a popular laboratory manual described that double stranded linear DNA fragments migrate approximately 10 % faster in TAE than in TBE (4), the small DNA fragments were not always following the rule. In this study, we employed TAE or TBE as an electrophoresis buffer and DNA size markers to compare the electrophoretic mobility as a practical bench mark for the benefit of a routine work in a laboratory.
MATERIALS AND METHODS
The DNA size markers (100 ng) , lambda DNA/HindIII fragments or phiX174 RF DNA /HaeIII fragments (New England BioLabs) were mixed with gel-loading buffer (6x buffer; 0.25% bromophenol blue, 0.25% xylene cyanol FF, 15% Ficoll), then loaded into the slots of the agarose (Ultra PURE agarose, GIBCOBRL) gel. A minigel (52 x 60 mm) submerged in running buffer, 0.5 x TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA pH 8.0) or 0.5 x TAE (20 mM Tris acetate, 1 mM EDTA pH 8.0) stored in an ultra mini chamber system (MupidTM-21, Cosmo-Bio, Japan). The gel was run in the absence of ethidium bromide, and stained after electrophoresis was completed in selected buffer containing ethidium bromide (0.5 mg/ml) for 20 min at room temperature.
RESULTS AND DISCUSSION
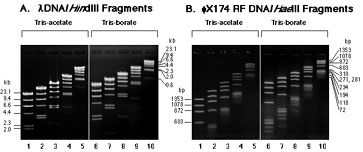
We analyzed two kinds of DNA size markers on the different concentrations of agarose gels containing TAE or
TBE buffers. lambdaDNA/HindIII fragments (23.1 to 0.6-kb) were analyzed on the 0.8 % agarose gels. phiX174
RF DNA /HaeIII fragments (1353 to 72-bp) are analyzed on 2.0 % agarose gels. It was clear that relatively
large DNA fragments (>2-kb on the 0.8 % agarose gel and >300 bp on 2.0 % agarose gel) were well separated on
the gels in TAE buffer than TBE buffer (Fig. 1). Conversely, we encountered the poor separation of smaller (<2-kb
on 0.8 % agarose gel and <300-bp on 2.0 % agarose gel) in TAE buffer. The smaller DNA fragments were more clearly
separated in TBE buffer (Fig. 1. lanes 6-10) than TAE buffer on each gel concentration (Fig. 1. lanes 1-5). Large
DNA fragments (23.1 to 2-kb) migrated faster in TAE than in TBE buffer on 0.8 % agarose gels, and small fragments
(2 to 0.6-kb) migrated faster in TBE than in TAE buffer. The crossing point on the graph dropped on 2-kb in length
(Fig. 2. A). Relatively large DNA fragments (1353 to 300-bp) migrated faster in TAE than in TBE buffer. The crossing
point on the graph showed 300-bp in length (Fig. 2. B).
In conclusion, TBE buffer was effective to obtain high resolution of smaller DNA fragments (300-bp), and TAE buffer
was useful for larger DNA fragments (<2.0-kb) in the mini chamber system (MupidTM-21). The conventional buffer,
TAE, is not always the best choice for gel electrophoresis. Considering the fragment size of DNA, an adequate
electrophoretic condition has to be chosen , i.e., TAE buffer is for larger DNAs than 2-kb in 0.8 % agarose,
and TBE for smaller DNAs than 300-bp in 2.0 % agarose. Either buffer, TBE or TAE, is applicable for the middle
size the DNA fragments ranging from 300-bp to 20-kb. The relative mobility varied according to the size of DNA
fragments, the concentration of agarose, and the composition of electrophoresis buffer. Although the DNA fragments
were generally believed to migrate faster in TAE than in TBE, the smaller fragments were not always the case.
They could migrate faster in TBE than in TAE. ¡ÈThis was sometime a paradox,
but now the time gives it proof¡É(5). Very recently, Stellwagen and colleagues have
given a possible explanation to show the paradox (6, 7). The higher mobility of small fragments in TBE may be
attributed to the formation of nonspecific, highly charged deoxyribose-borate complexes (7).
LEGENDS FOR FIGURES

Fig. 1. DNA electrophoresis with Tris-acetate buffer (TAE) or Tris-borate (TBE) on agarose gels. A. lambdaDNA/HindIII Fragments (100 ng) were loaded on 0.8% agarose gel and separated by electrophoresis in Tris-acetate buffer (lane 1-5) or Tris-borate buffer (lane 6-10). Running time for 10 min (lanes 5, 10), 20 min (lames 4, 9), 30 min (lames 3, 8), 40 min (lanes 2, 7), 50 min (lanes 1, 6). B. phi174/HaeIII RF DNA Fragments 100 ng were loaded on 2.0% agarose gel and separated by electrophoresis in Tris-acetate buffer (lane 1-5) or Tris-borate buffer (lane 6-10). Running time described in A.
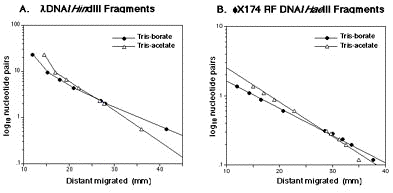
Fig. 2. Mobility of the DNA fragments on agarose gel in Tris-acetate buffer (TAE) or Tris-borate (TBE). A. phiDNA/HindIII Fragments 100 ng were loaded on 0.8% agarose gel and separated by electrophoresis in Tris-acetate buffer ( ) or Tris-borate buffer (¡ü). B. phi174 RF DNA/Hae III Fragments (100 ng) were loaded on 2.0% agarose gel and separated by electrophoresis in Tris-acetate buffer ( ) or Tris-borate buffer (¡ü). Running time described in A.
REFERENCES
1. Loening, U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem. J. 1967 ; 102: 251-257.
2. Ogden, Richard C. and Deborah A. Adams. In: Berger, Shelby L. and Alan R. Kimmel eds. Guide to Molecular Cloning Techniques (Methods in Enzymology Vol. 152) Academic Press Inc. Orlando, Florida. 1987; 75-78.
3. Dingman, C. W. and A. C. Peacock. Analytical studies on nuclear ribonucleic acid using polyacrylamide gel. Biochemistry 1968; 7: 659-668.
4. Sambrook, J., Fritsch, E. F. and Maniatis T. Molecular cloning - a laboratory manual 2nd ed. Cold Spring Harbor Lab., Cold Spring Harbor, New York. 1989: 6.4-6.5.
5. Shakespeare, W. In: HAMLET Act3 Scene 1. The Oxford Shakespeare. the complete works. Oxford University Press Inc., New York 1988: 670.
6. Stellwagen, N.C. Apparent pore size of polyacrylamide gels: comparison of gels cast and run in tris-acetate-EDTA and tris-borate-EDTA buffers. Electrophoresis 1998; 19: 1542-1547.
7. Struz, K. and Stellwagen, N.C. Do DNA gel electrophoretic mobilities extrapolate to the free-solution mobility of DNA at zero gel concentration? Electrophoresis 1998; 19: 635-642.
Key words:
Tris-borate, TBE, Tris-acetate, TAE, running buffer, agarose gel electrophoresis
|
Medical School |